Breast Cancer Hackathon Challenge 2023
Anticipated in Spring 2023, Komen is excited to announce the Breast Cancer Hackathon Challenge!
The goal of the Hackathon is to bring together creative individuals address “challenge questions” that represent key unmet needs in the field of breast cancer by leveraging big data in innovative ways.
The Breast Cancer Hackathon Challenge is a partnership between Susan G. Komen, the University of Texas Southwestern (UTSW) Lyda Hill Department of Bioinformatics, and the UTSW Harold C. Simmons Comprehensive Cancer Center with the generous support of Lyda Hill Philanthropies.
Careers in Computational Biology
Date: Friday, March 3rd, 2023
Time: 1 – 4 PM
Location:
Lyda Hill Department of Bioinformatics, J9 Building
5323 Harry Hines Blvd., Dallas
Speaker & Career Mentor Bios
the Breast Cancer Hackathon Challenge 2023 kicks off from 4 PM onwards.
The goal of the Hackathon is to bring together creative individuals address “challenge questions” that represent key unmet needs in the field of breast cancer by leveraging big data in innovative ways.
The Breast Cancer Hackathon Challenge is a partnership between Susan G. Komen, the University of Texas Southwestern (UTSW) Lyda Hill Department of Bioinformatics, and the UTSW Harold C. Simmons Comprehensive Cancer Center with the generous support of Lyda Hill Philanthropies.
Where: UTSW Campus, Dallas, TX
When: March 3-5, 2023
PROJECT 1
Predict and visualize combined functional effects of germline and somatic alterations in breast cancer
Cancer research focuses on a few dozen “cancer driver” genes and tends to ignore the thousands of other germline and somatic alterations present in every cancer, which are assumed to be functionally irrelevant “passengers”. This view is too simplistic. Every cancer has a unique clinical behavior the same way as every person has a unique face. The uniqueness of one’s face, and organs, is due to the combined effect of thousands of polymorphisms that one was born with compounded by somatic epigenetic changes acquired during aging. The challenge here is to develop a tool that captures the combined biologic effects of all germline and somatic alterations within a particular breast cancer.
What contributions can individuals make with working on this project?
‘One size fits all’ shouldn’t be how we approach breast cancer treatment. Current pathway models are complex but do not account for one's individuality. Help us bring precision medicine to all people.
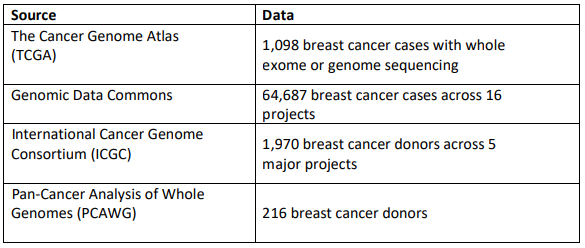
What data will we provide to work on this project?
The following are examples of datasets that could be used for working on Project 1.
PROJECT 2
Define and compare metabolic states of cell types found in the breast tumor microenvironment
One approach to target advanced breast cancer is to identify and therapeutically target the metabolic vulnerabilities of key players in the tumor microenvironment (cancer cells, fibroblasts, immune cells, etc.). However, this has been very difficult to study due to technological limitations. With the recent advent of big data in human breast cancer, especially single cell RNA-seq (scRNAseq), the goal is to infer the comprehensive metabolic activities within and among major cell types in the tumor microenvironment using scRNA-seq datasets of human breast cancer.
What contributions can individuals make with working on this project?
It has been known for a long time that ‘cancer’ is not just a matter of cancer cells. Interaction with other cells often determine the onset and progression of cancer. Distinguishing the contribution of different cell types is limited when leveraging whole tumor data.
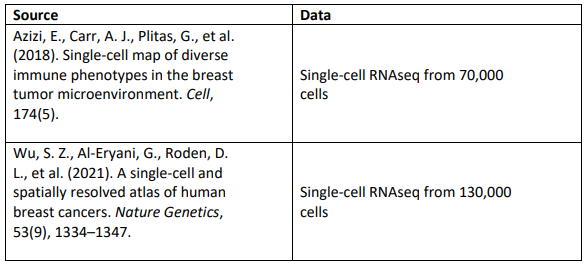
What data will we provide to work on this project?
The following are examples of datasets that could be used for working on Project 2.
PROJECT 3
Identify and prioritize personalized drug combinations based on the genomic landscape of breast cancer
Breast cancer cells exploit multiple pathways to evade the selective pressure of single drugs, promoting therapeutic resistance and clinical relapse. More rational identification of new targets in breast cancer for combination drug regimens is an essential next step in providing long-term clinical benefits. The goal is to accelerate the discovery of combination therapies through integrative, systematic network-based identification of co-occurring genomic alterations in breast cancer patients. The accumulation of omics data from breast cancer increases the feasibility and chance of success of computational analyses to identify synergistic interactions for combination therapy, which has been technically challenging. Therefore, the development of efficient in silico screens and prioritization of co-targetable pathways will be critical, enabling more powerful combinatorial therapeutics for breast cancer.
What contributions can individuals make with working on this project?
Oncologists regularly get to expand their toolbox with new drugs, and one only hopes to determine the optimal treatments available to patients. Identification of combination therapies is limited by the number of possible drug combinations and genomic alterations.
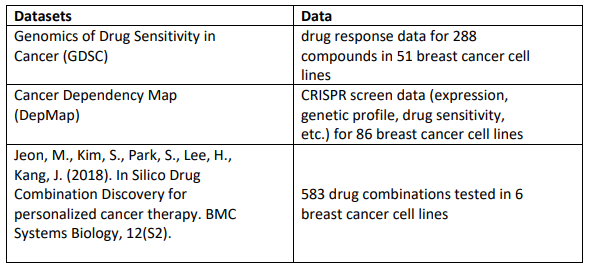
What data will we provide to work on this project?
The following are examples of datasets that could be useful for working on Project 3.